Drop-seqDroplet Based scRNA-Seq Methods
Drop-seq (Macosko et al., 2015**) is one of the popular of the scRNA-seq methodologies. The strategy, using droplet microfluidics, gives access to the transcriptomes of thousands of single cells and enables researchers to understand tissues at the level of individual cells.The popularity of Drop-seq is owing to several benefits:
● High throughput – 5,000 – 10,000 single cell transcriptomes per run
● High capture rate of around 11% of transcriptomes of cells
● Low doublet rate of about 5%
● Simplicity
● Commercial availability of the barcoded beads
● Relatively low costThe methodology involves encapsulating single cells with single barcoded beads in nanoliter-sized droplets. The barcoded oligo bead library is constructed such that each bead has a unique DNA barcode sequence, but within a bead, the thousands of copies of oligo all contain an identical barcode sequence. The 3′ end of the oligo has a poly(dT) stretch that captures mRNA and primes reverse transcription. Once encapsulated, the cell is broken open and the mRNA is captured on the bead, resulting in STAMPs (Single-cell Transcriptomes Attached to MicroParticles). The RNA is converted to DNA, amplified and sequenced.Core Principles of Drop-seq
● Encapsulation of single cells with DNA-barcoded microbeads
● The use of a microfluidic device/chip to create nanoliter-scale aqueous droplets formed by precisely combining aqueous and oil flows
● The use of molecular barcoding beads to distinguish the cell-of-origin of each mRNA
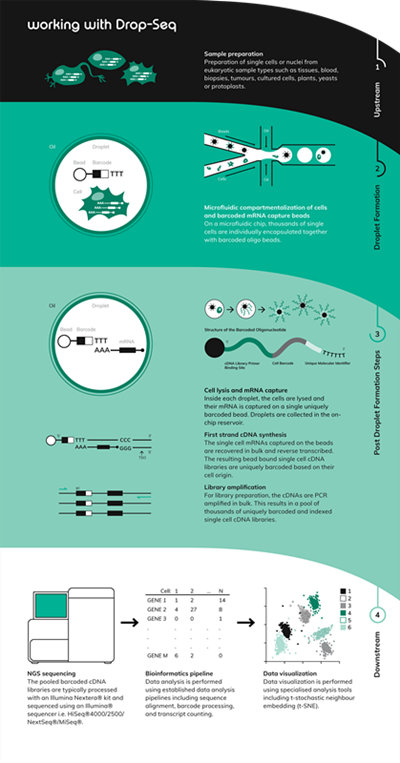
• Reverse transcription added post encapsulationDolomite Bio’s Nadia instrument seamlessly adopts the Drop-seq protocol. However, with the flexible and open nature of the system, there is also potential for alternative droplet-based methods to be adopted including inDrops, Hi-SCL, Dronc-seq, Perturb-seq and CROP-seq
Dolomite Bio’s Drop-seq Workflow